Introduction
Disorder is often called the hidden disability because our behavior and thinking may have very serious problems, and yet disorder patients always have no obvious physical changes. That’s the reason why this hidden disability is easily ignored and misunderstood.
Also, disorder is not an intellectual disability. Intelligence is usually not affected, although there are often cognitive changes, such as memory, attention, and attention. Here we give the precise definition of disorder, a disruption of the disease to the normal or regular functions in the body or a part of the body\(^[1]\).
Moreover, disorders can be classified into the following areas:
- Mental
- Physical
- Genetic
- Emotional
- Behavioural
- Structural
Following, we will focus on mental disorder.
A mental disorder is usually called a mental illness or a Schizophrenia. Different with the normal disease, mental disorder is a behavioral or mental pattern that causes significant distress or impairment of personal functioning. Such features may be persistent, relapsing and remitting, or occur as a single episode. This disorder disease may be diagnosed by a mental health professional.
How to diagnose the mental disorders?
Thorough mental examination, physical and neurological examination, laboratory examination, brain imaging examination and neuropsychological assessment, the psychiatrist can only make a preliminary judgment on the patient’s current mental state, and then the psychiatrist need to combine the complete medical history data, such as the personal life history and the related social and psychological factors, so that they can finally make the current diagnosis by analysing and summarizing the above informations.
Goal
To assist the psychiatrist in diagnosing and simplify the consultation process, we try to classify the mental disorder patients by analysing the MR brain images.
Data
Brain MR “T1” Images
- Source: Dr. Albert C Yang and Jean Lee
- Cases Number:
- Total: 400
- Healthy: 200
- Schizophrenia: 200
- Type: NIFTI file
- Spacing: 1 mm \(\times\) 1 mm \(\times\) 1 mm
- Size: 3D but not fiexd (I will cut into the smallest size of those images)
- Dataset:
- Training Set: 280 (70%)
- Validation Set: 40 (10%)
- Testing Set: 80 (20%)
Devices
- CPU: Intel(R) Core(TM) i9-7900X CPU @ 3.30GHz \(\times\) 20 Cores
- GPU: GeForce RTX 2080 \(\times\) 4 Cores
Methods
- Method 1 (Use CNN model):
- Cut all MR images to the same size since input layer only accepts the same size
- Train the CNN model and predict
- Tesorflow (v.1.13.1) Model Graph (display by Tensorboard):
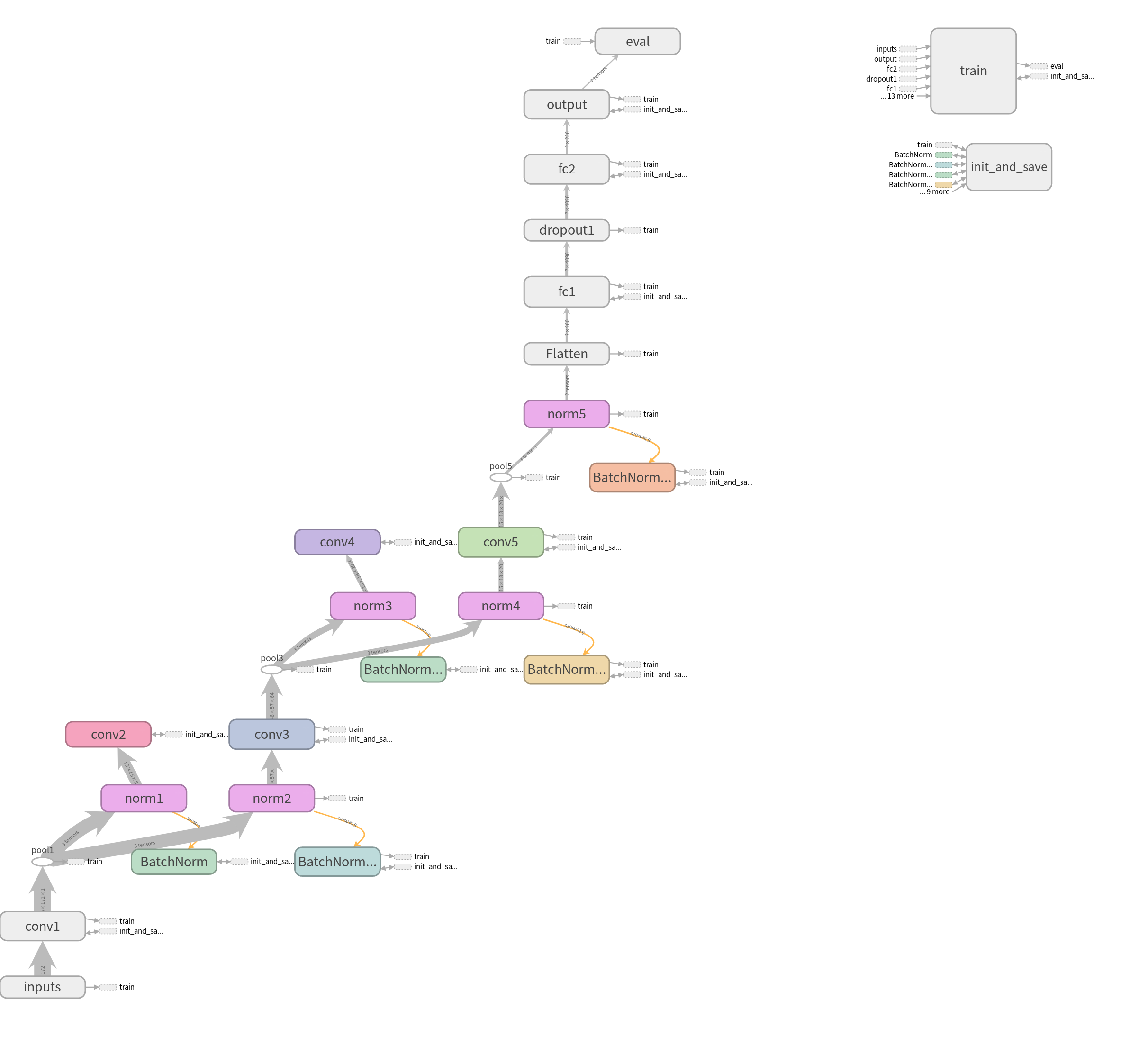
Fig1. CNN model
- Method 2 (Use Radiomics):
- Construct brain mask:
nipy.labs.mask.compute_mask_files \(\rightarrow\) scipy.ndimage.morphology.binary_fill_holes - Compute Radiomics features (v2.1.0)
- Compute ICC to extract useful Radiomics features
- Design classification model
- Construct brain mask:
- Method 3 (Use CNN model with segmentation MR images):
- Segment the white matter (MW) from MR brain images
- Normalize to 0 ~ 1024.
- Reduce one convolution since segmentation images are smaller than original size
- Train the CNN model and then predict
- Method 4 (Try to augment data):
- Use numpy or scipy package to rotation, translation, reflection and zoom all brain MR images
- Cut all MR images and augmentation MR images to the same size
- Train the CNN model and then predict
- Augment the segmentation data (WM images)
- Train the CNN model and then predict again
Results
- Method 1 (Use CNN model):
1. Learning Rate: \(1\times10^{-5}\), Batch Size: 10, Epoch: 100
Training Results Figure:
a. Accuracy:
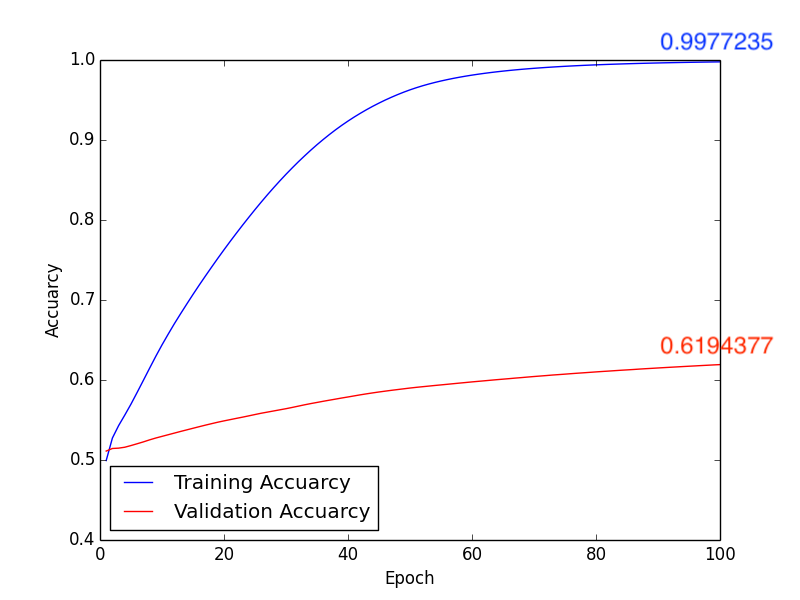
Fig2. Accuracy
b. Loss:
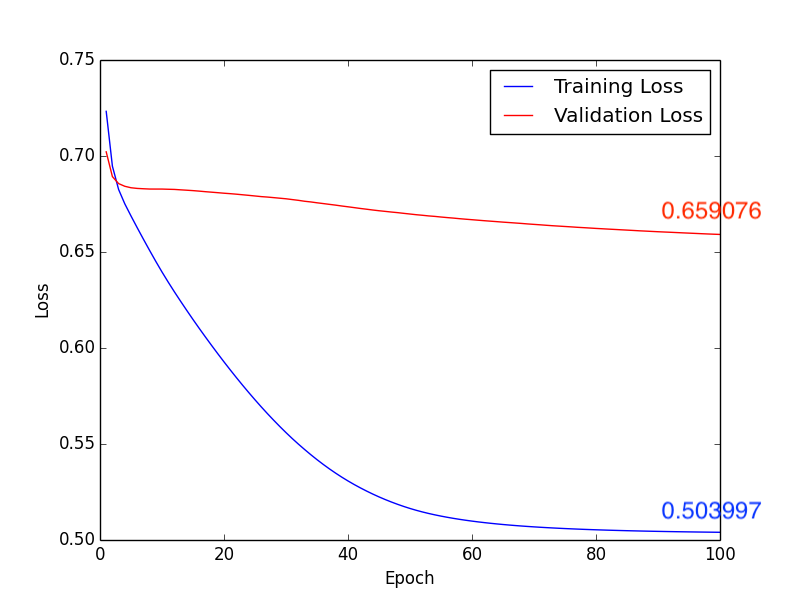
Fig3. Loss
Testing Accuracy: 0.625
Confusion Matrix:
| Actual | Predict | |
|---|---|---|
| Negative | Positive | |
| Negative | 33 | 10 |
| Positive | 20 | 17 |
Form1. Confusion matrix
2. Learning Rate: \(1\times10^{-4}\), Batch Size: 10, Epoch: 100
Training Results Figure:
a. Accuracy:
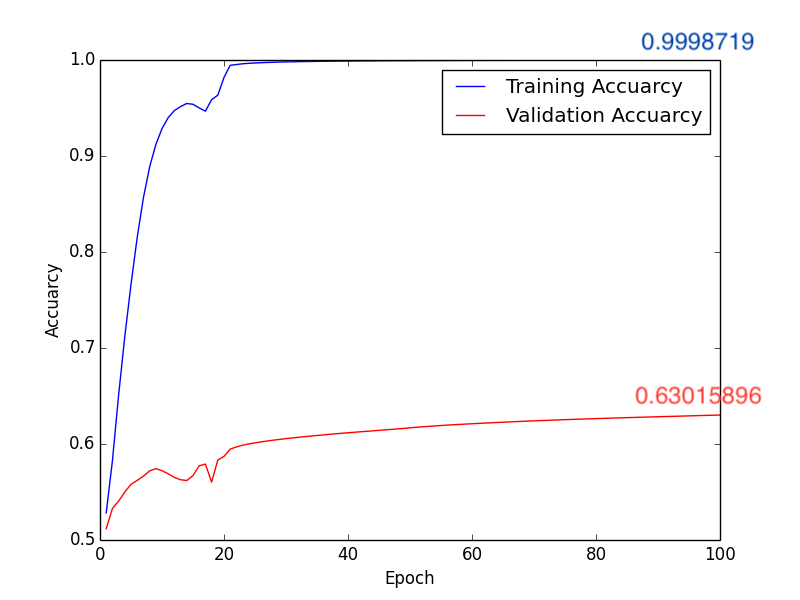
Fig4. Accuracy
b. Loss:
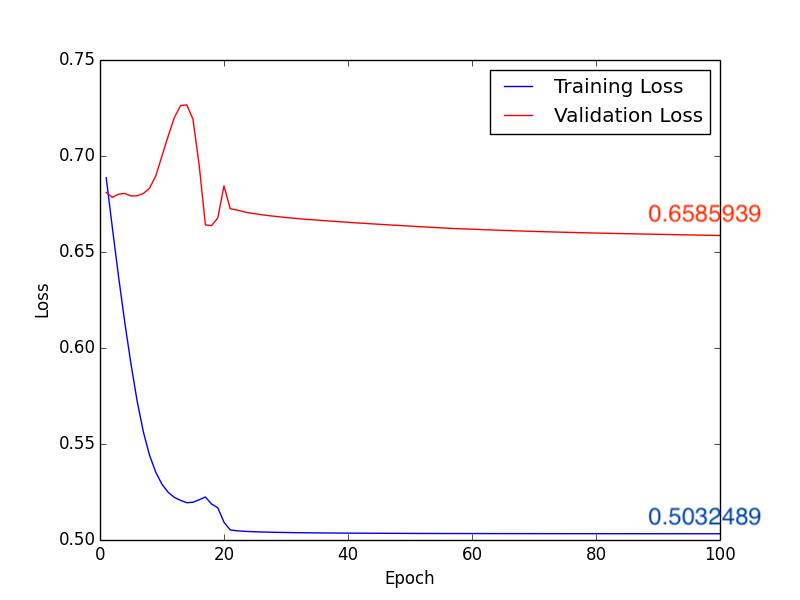
Fig5. Loss
Testing Accuracy: 0.6625
Confusion Matrix:
| Actual | Predict | |
|---|---|---|
| Negative | Positive | |
| Negative | 33 | 10 |
| Positive | 17 | 20 |
Form2. Confusion matrix
- Method 2 (Use Radiomics):
1. Construct brain mask:
a. Original brain images:
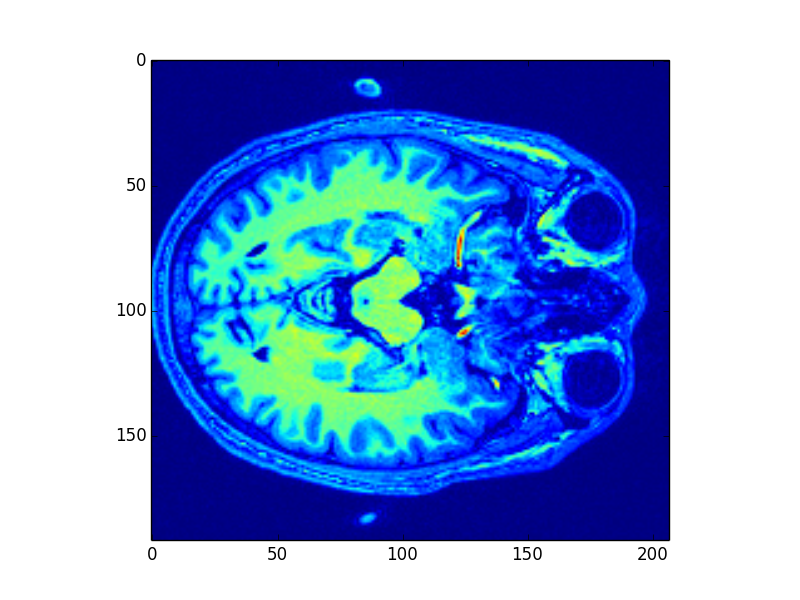
Fig6. Slice of the brain MR image
b. Generate brain mask:
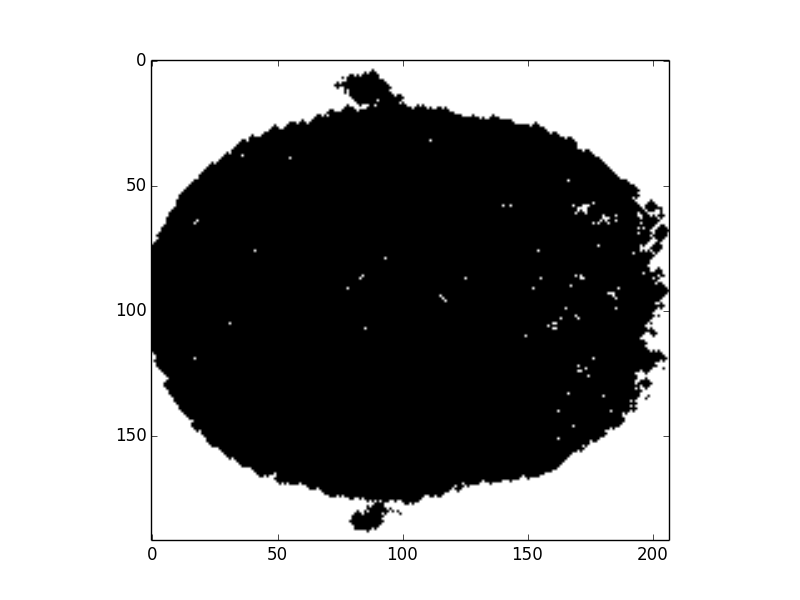
Fig7. Preliminary brain mask
c. Fill the holes:
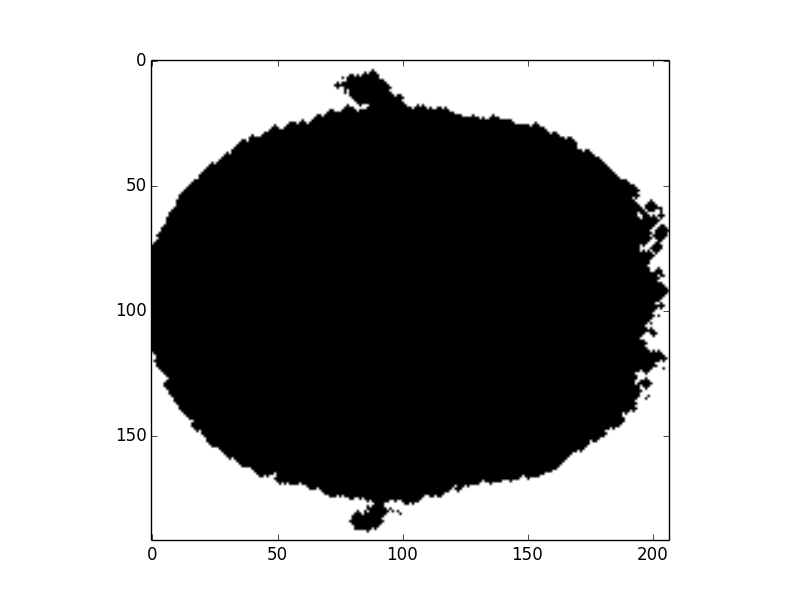
Fig8. Final brain mask
2. Extract Radiomics features:
a. Use defalut Radiomics setting and total meaningful features is 107
i. First Order Features
ii. Gray Level Co-occurrence Matrix Features (GLCM)
iii. Gray Level Dependence Matrix Features (GLDM)
iv. Gray Level Run Length Matrix Features (GLRLM)
v. Gray Level Size Zone Matrix Features (GLSZM)
vi. Neighbouring Gray Tone Difference Matrix Features (NGTDM)
3. Use ICC to extract useful Radiomics features:
a. Use function “rpy2.robjects.packages.importr(“ICC”)” to compute ICC values.
b. ICC values:
We get very pool results, since all ICC values are less than 0.4 (poor correlation). You can check the more detail Radiomics results in GitHub.
4. Design model:
ICC results are very weak so that we cannot to design robust model to classify.
- Method 3 (Use CNN model with segmentation MR images):
1. Segment the white matter
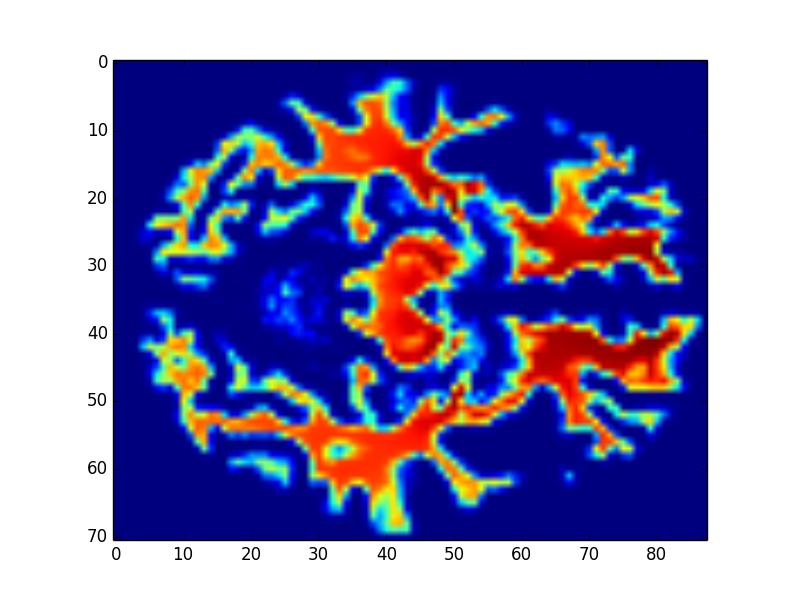
Fig9. White matter
2. Use linear normalize original value of the white matter to 0 ~ 1024
3. Learning Rate: \(1 \times 10^{-5}\), Batch Size: 10, Epoch: 100
Training Results Figure:
a. Accuracy:
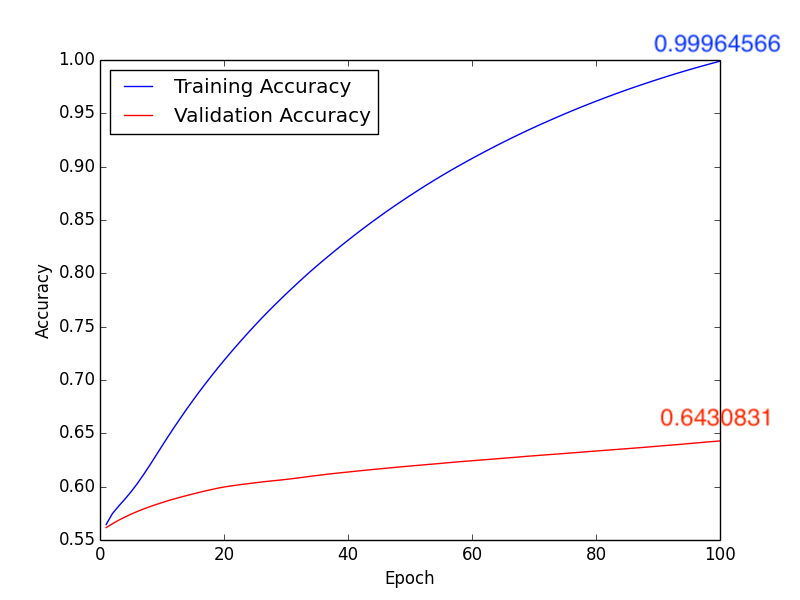
Fig10. Accuracy
b. Loss:
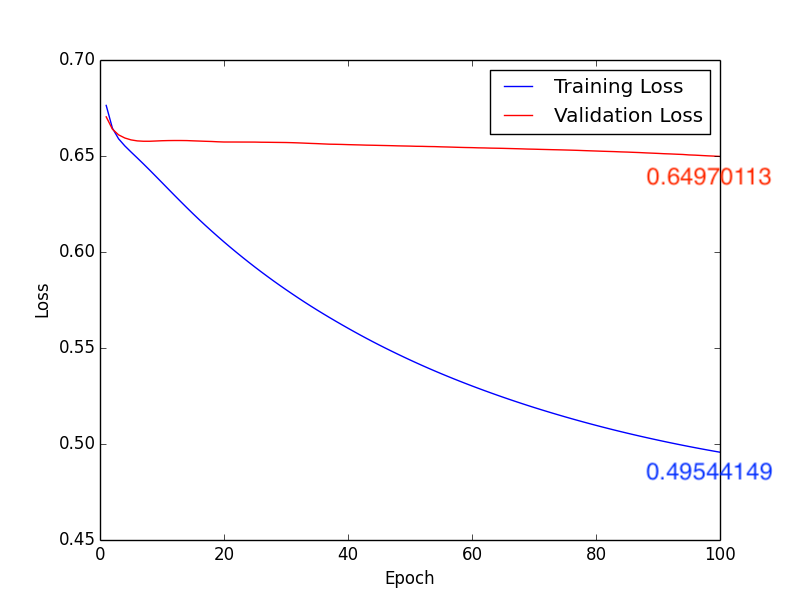
Fig11. Loss
Testing Accuracy: 0.7125
Confusion Matrix:
| Actual | Predict | |
|---|---|---|
| Negative | Positive | |
| Negative | 36 | 7 |
| Positive | 16 | 21 |
Form3. Confusion matrix
4. Learning Rate: \(1 \times 10^{-4}\), Batch Size: 10, Epoch: 100
Training Results Figure:
a. Accuracy:
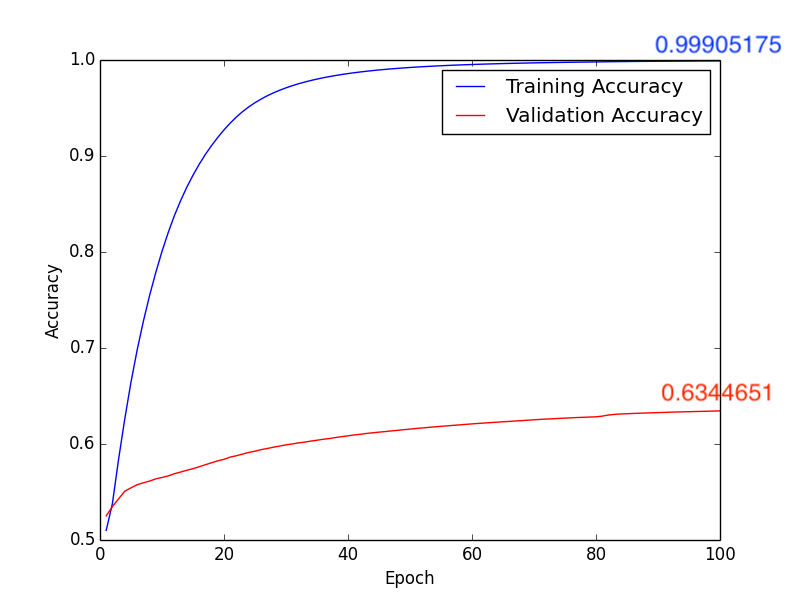
Fig12. Accuracy
b. Loss:
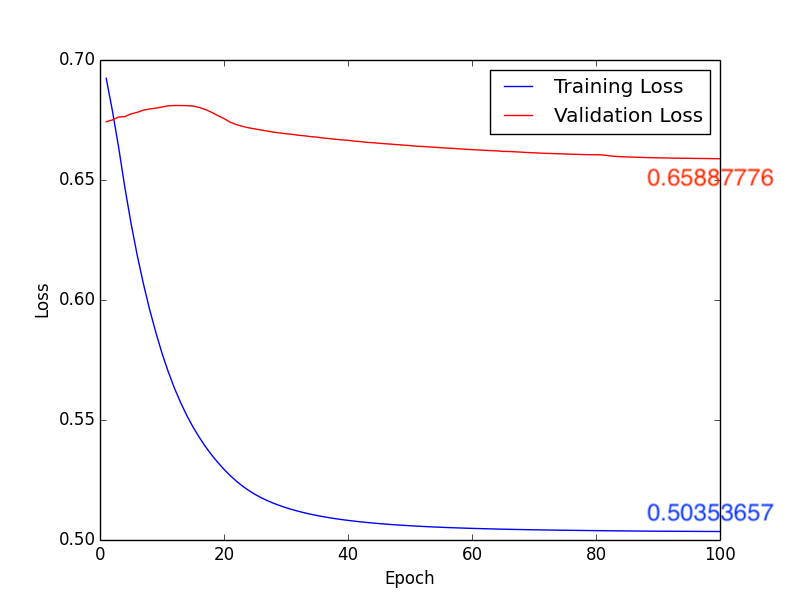
Fig13. Loss
Testing Accuracy: 0.65
Confusion Matrix:
| Actual | Predict | |
|---|---|---|
| Negative | Positive | |
| Negative | 32 | 11 |
| Positive | 17 | 20 |
Form4. Confusion matrix
- Method 4 (Try to augment data):
- The training, validation and testing accuracy will reduce to 0.5
- The results are extremely worse so that we will not upload this code in GitHub
- The reasons why we get this weak performance will be explained in the following conculsions
Conclusions
In this classification task, we try to use deep learning model and Radiomics to achieve the goal. First method, after directly constructing the CNN model (Fig1.) to train and predict, and adjusting some hyperparameters, we get the following results, compare with Fig2. and Fig4., we can find that the higher accuracy occurs when using \(1 \times 10^{-4}\) learning rate. Moreover, in the Form2., the number of FP is lower than Form1. but the FN is equivalent. It seems that lower FN is more important than lower FP, because if we judge the schizophrenia patient as the healthy person, the patient may not be able to receive treatment immediately, or even worsen the condition. So it’s necessary to come up with other methods to imporve the performance of method 1.
In the method 2, we use Radiomics to classify the brain MR images. To let py-Radiomics working, we need to construct the brain mask first (Fig6. and Fig7.). Since Radiomics will not work very well with the hollow mask, we also need to fill all holes in the mask (Fig8.). After getting 107 Radiomics features, the vital step is to select the useful features by ICC. Unfortunately, all ICC values are lower than 0.4, so this meathod is unacceptable.
About method 3, according to pervious sutdies\(^{[2], [3]}\), we know that the white matter is highly realtive to Schizophrenia. Isteading of using whole brain MR images, we train the CNN model with the segmentation images of the white matter (Fig9.). Compare with Fig10. ~ Fig13., we get the higher accuracy by using learning rate \(10 \times 10^{-4}\). In addition, comparing with Form1. ~ Form4., method 3 with \(1 \times 10^{-4}\) leanring rate will have the higher accuracy, lower FP and “lower FN”.
Final method, we try to augment the dataset since we notice that our dataset might be shortage. Because the human brain structure is very complex and explicit, we can not use rotation rotation, translation, reflection and zoom to augment our dataset. That’s the reason why these weak performance.
Future works
- Find out the reason why Radiomics doesn’t work very well and then fix this problem
- Solve the problem of data shortage and train the model again
- Improve the validation and testing accuracy of the recent model to 0.8
References
- World Health Organization, Department of Mental Health and Substance Dependence. Investing in MENTAL HEALTH. 2003.
- Andrew M. McIntosh, Susana Muñoz Maniega, G. Katherine S. Lymer and et al. White Matter Tractography in Bipolar Disorder and Schizophrenia. Dec 2008.
- Kenneth L. Davis, Daniel G. Stewart, Joseph I. Friedman and et al. White Matter Changes in Schizophrenia: Evidence for Myelin-Related Dysfunction. May 2003.
- Mahbub Hussain, Jordan J. Bird and Diego R. Faria. A Study on CNN Transfer Learning for Image Classification. June 2018.
- Paras Lakhani, Daniel L. Gray, Carl R. Pett, Paul Nagy and George Shih. Hello World Deep Learning in Medical Imaging. May 2018.
- Muhammad Imran Razzak, Saeeda Naz and Ahmad Zaib. Deep Learning for Medical Image Processing: Overview, Challenges and Future.
- Despina Kontos, Ronald M. Summers and Maryellen Giger. Radiomics and Deep Learning. J. Med. Imag. 2018.
- Stefania Rizzo, Francesca Botta, Sara Raimondi, Daniela Origgi, Cristiana Fanciullo, Alessio Giuseppe Morganti and Massimo Bellomi. Radiomics: the facts and the challenges of image analysis. Dec 2018.
- Tensorflow Guide. (“https://www.tensorflow.org/guide”)
- Py-Radiomics. (“http://www.radiomics.io/pyradiomics.html”)
Acknowledgements
I would like to thank Dr. Albert C Yang and Jean Lee who provided all brain MR images and gave me guidance and advice.

Leave a Comment
Your email address will not be published. Required fields are marked *